Locomotion visualizations
Select & Summarize
play_loco <- home_visit_df %>%
dplyr::select(
.,
age_group,
child_sex,
language_child,
site_id,
subject_number,
locomotor_milestones.who_walk.who_walk_onset_mo,
locomotor_milestones.k_walk.k_walk_onset_mo,
locomotor_milestones.crawl_onset.crawl_onset_mo
) %>%
dplyr::rename(
.,
walk_mos_who = locomotor_milestones.who_walk.who_walk_onset_mo,
walk_mos_kea = locomotor_milestones.k_walk.k_walk_onset_mo,
crawl_mos = locomotor_milestones.crawl_onset.crawl_onset_mo
) %>%
dplyr::mutate(
.,
walk_mos_who = as.numeric(walk_mos_who),
walk_mos_kea = as.numeric(walk_mos_kea),
crawl_mos = as.numeric(crawl_mos)
)
xtabs(formula = ~ child_sex + age_group, data = play_loco)## age_group
## child_sex 12mo 18mo 24mo
## female 144 129 109
## male 122 149 113Check for anomalous values
crawl_mos_min <- 4
walk_mos_min <- 6Anomalous crawling onset
play_loco %>%
dplyr::select(., site_id, subject_number, crawl_mos) %>%
dplyr::filter(., crawl_mos < crawl_mos_min) %>%
knitr::kable(format = 'html') | site_id | subject_number | crawl_mos |
|---|---|---|
| CHOPH | 003 | 3.72 |
| PRINU | 020 | -5.69 |
Crawl onset
play_loco %>%
dplyr::filter(., crawl_mos > crawl_mos_min, !is.na(crawl_mos)) %>%
ggplot(.) +
aes(crawl_mos, fill = child_sex) +
geom_histogram(bins = 12) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())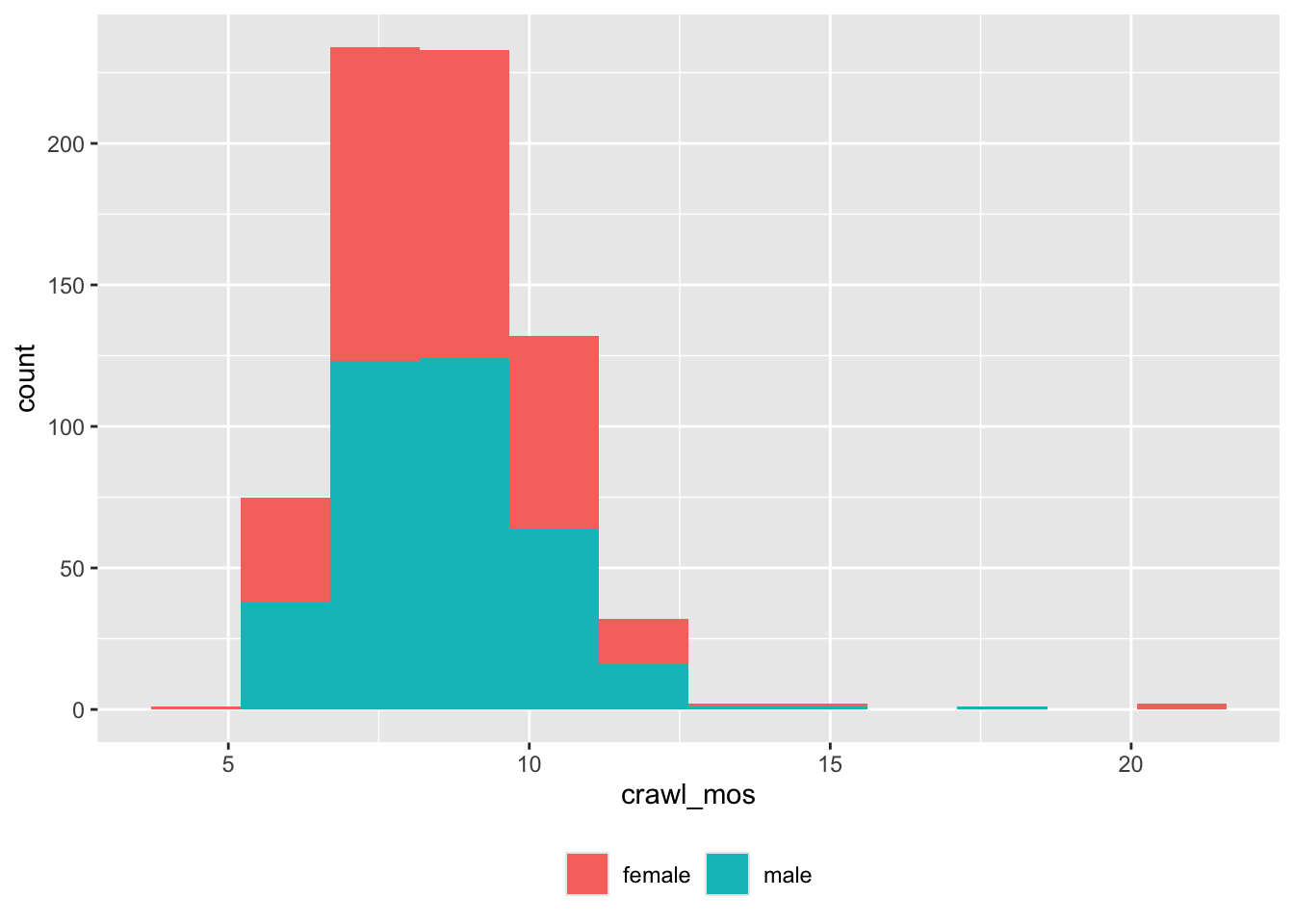
Figure 6: Age of crawling onset (mos) by sex
Walk onset
play_loco %>%
dplyr::filter(., walk_mos_kea > walk_mos_min, !is.na(walk_mos_kea)) %>%
ggplot(.) +
aes(walk_mos_kea, fill = child_sex) +
theme(legend.position="bottom") +
geom_histogram(bins = 10)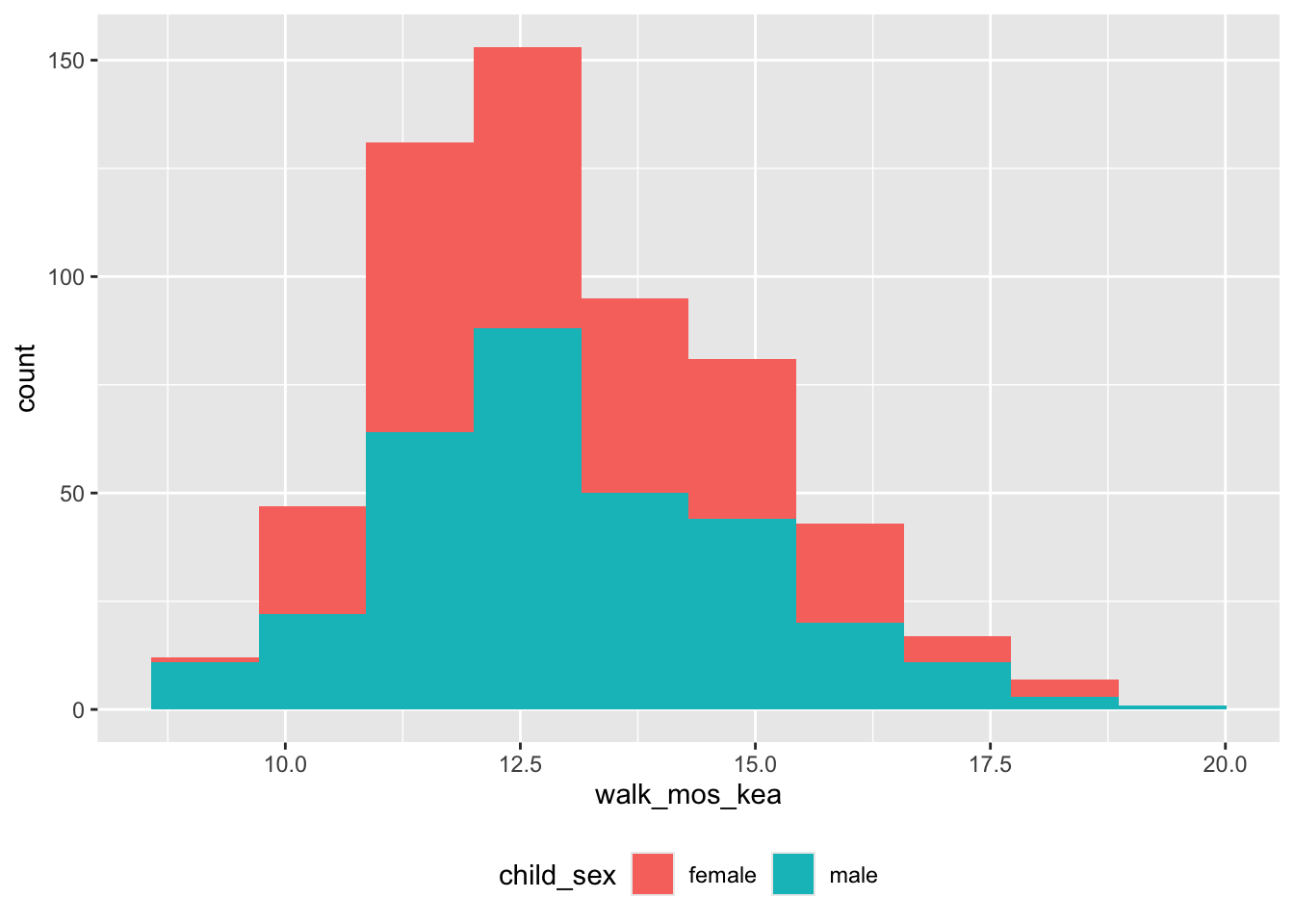
Figure 7: Age (mos) of walking onset (KEA criteria) by sex
play_loco %>%
dplyr::filter(., walk_mos_who > walk_mos_min, !is.na(walk_mos_who)) %>%
ggplot(.) +
aes(walk_mos_who, fill = child_sex) +
geom_histogram(bins=12) +
theme(legend.position="bottom") +
theme(legend.title = element_blank())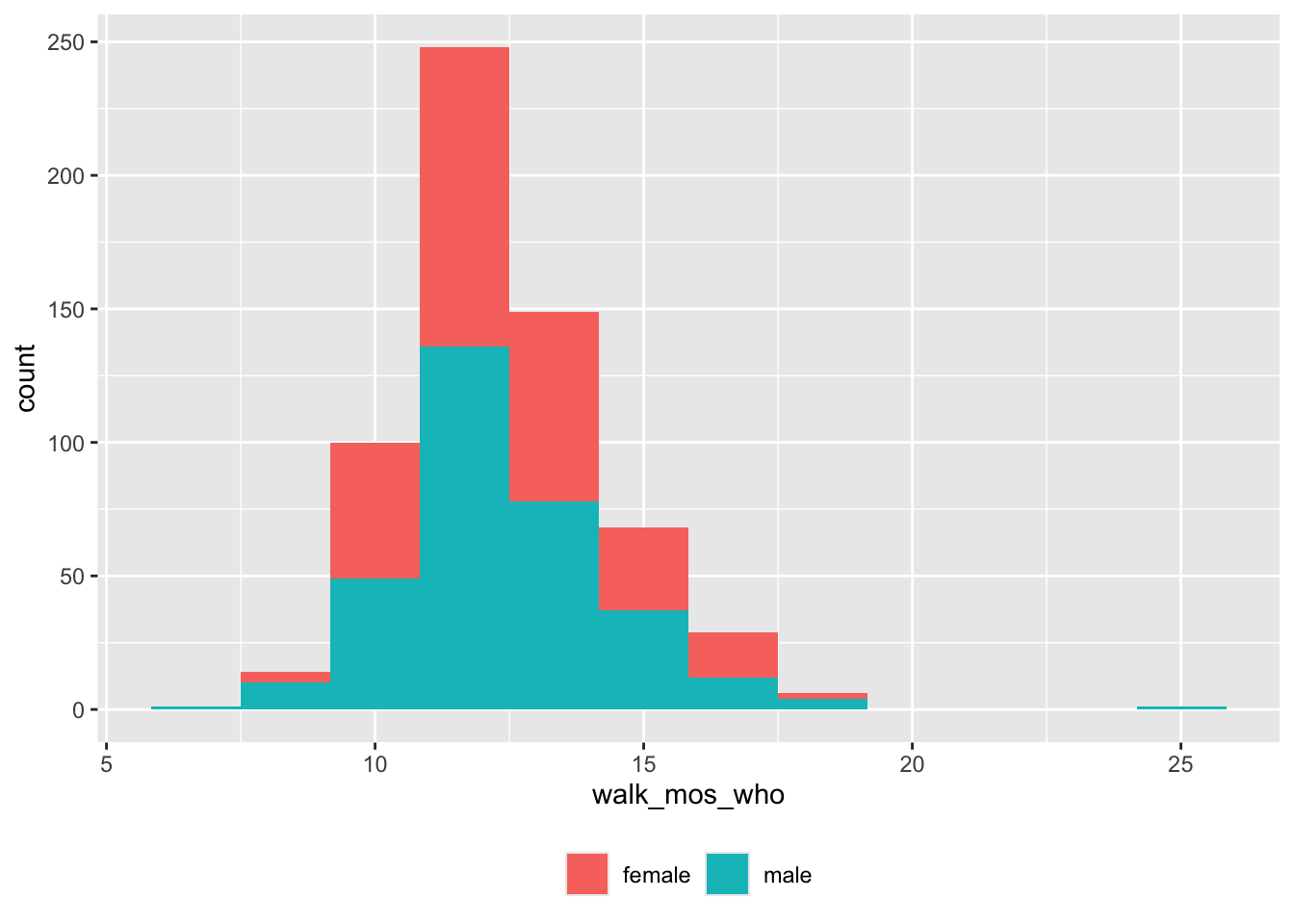
Figure 8: Age (mos) of walking onset (WHO criteria) by sex
play_loco %>%
dplyr::filter(., walk_mos_who > walk_mos_min, !is.na(walk_mos_who),
walk_mos_kea > walk_mos_min, !is.na(walk_mos_kea)) %>%
ggplot(.) +
aes(walk_mos_who, walk_mos_kea, color = child_sex) +
geom_point() +
geom_smooth(method = "lm") +
xlim(8, 18) +
ylim(8, 18) +
theme(legend.position = "bottom") +
theme(aspect.ratio = 1) +
theme(legend.title = element_blank()) -> walk_p
ggExtra::ggMarginal(
walk_p,
play_loco,
walk_mos_who,
walk_mos_kea,
type = "density",
margins = "both",
groupColour = TRUE,
groupFill = TRUE
)## `geom_smooth()` using formula = 'y ~ x'## Warning: Removed 9 rows containing non-finite outside the scale range
## (`stat_smooth()`).## `geom_smooth()` using formula = 'y ~ x'## Warning: Removed 9 rows containing non-finite outside the scale range
## (`stat_smooth()`).## Warning: Removed 9 rows containing missing values or values outside the
## scale range (`geom_point()`).## Warning: Removed 1 row containing missing values or values outside the
## scale range (`geom_smooth()`).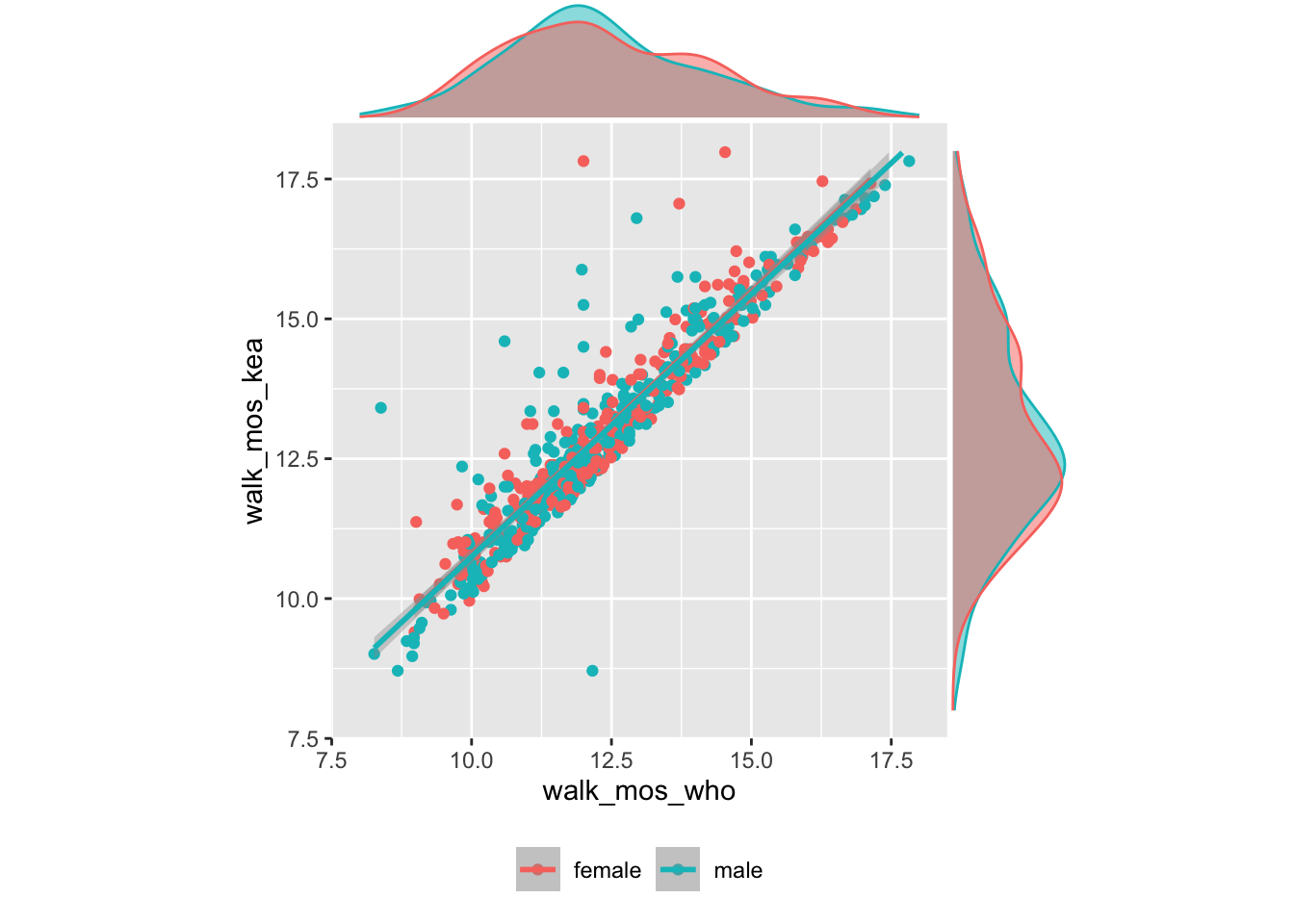
Figure 9: Walking onset by WHO vs. KEA criteria
play_loco %>%
dplyr::filter(., crawl_mos > crawl_mos_min, !is.na(crawl_mos),
walk_mos_kea > walk_mos_min, !is.na(walk_mos_kea)) %>%
ggplot(.) +
aes(crawl_mos, walk_mos_kea, color = child_sex) +
geom_point() +
geom_smooth(method = "lm") +
theme(legend.position = "bottom") +
theme(aspect.ratio = 1) +
theme(legend.title = element_blank()) -> walk_p
ggExtra::ggMarginal(
walk_p,
play_loco,
walk_mos_who,
walk_mos_kea,
type = "density",
margins = "both",
groupColour = TRUE,
groupFill = TRUE
)## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'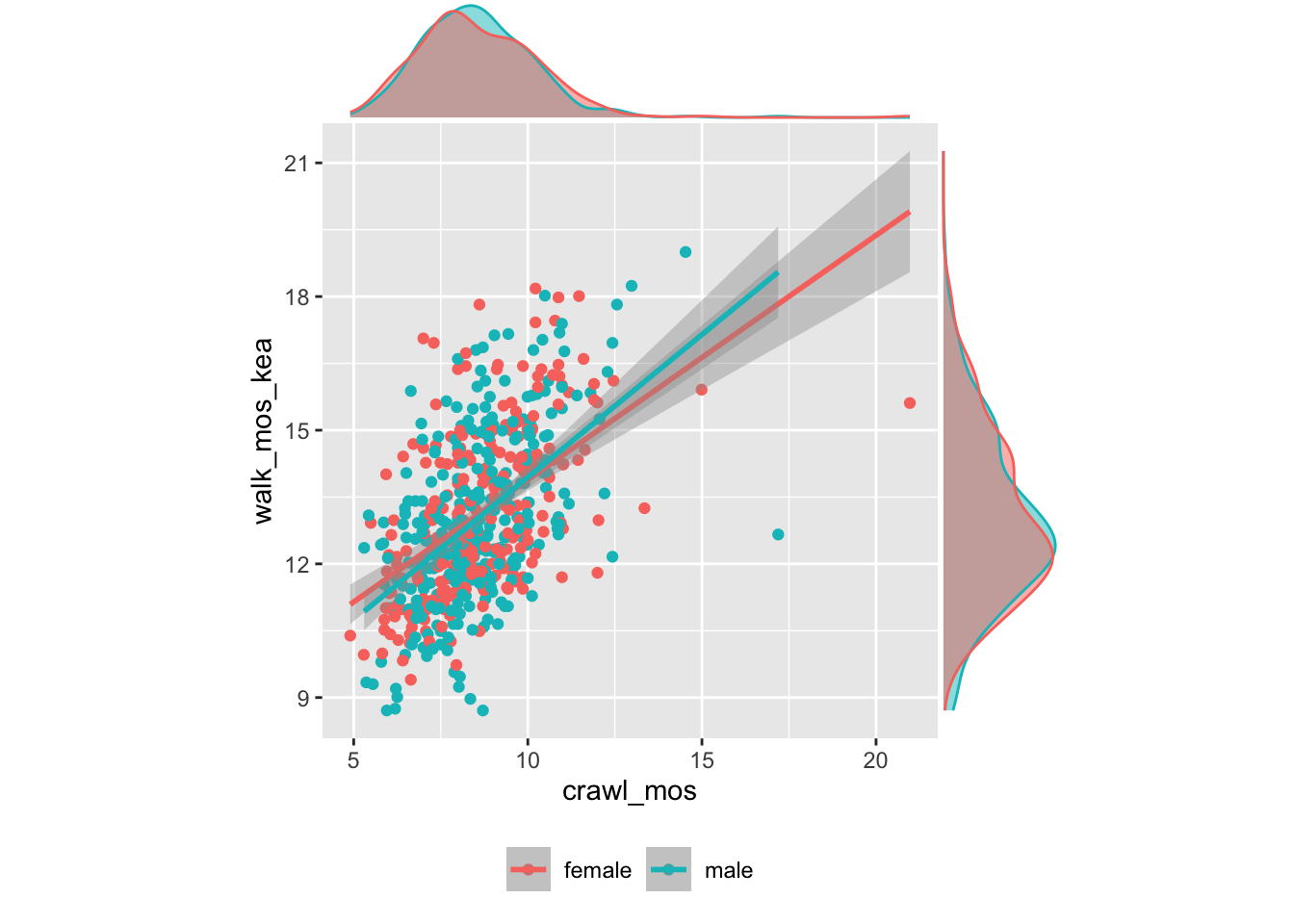
Figure 10: Walking onset vs. Crawling