Screening visualizations
These visualizations are intended as a way to test the integrity and utility of the data export and cleaning workflow.
Setup
screen_df <- readr::read_csv(file.path(here::here(), "data/csv/screening/agg/PLAY-screening-datab-latest.csv"))## Rows: 816 Columns: 68
## ── Column specification ─────────────────────────────────────────
## Delimiter: ","
## chr (55): session_name, session_release, participant_ID, par...
## dbl (9): session_id, vol_id, child_age_mos, child_birthage,...
## lgl (1): pilot_pilot
## dttm (1): submit_date
## date (2): session_date, participant_birthdate
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Dates & times
To calculate cumulative screening/recruiting calls by site, we have to add an index variable
Calls across time
df |>
dplyr::filter(!is.na(submit_date), !is.na(n_calls), !is.na(site_id)) %>%
ggplot() +
aes(submit_date, n_calls, color = site_id) +
geom_point()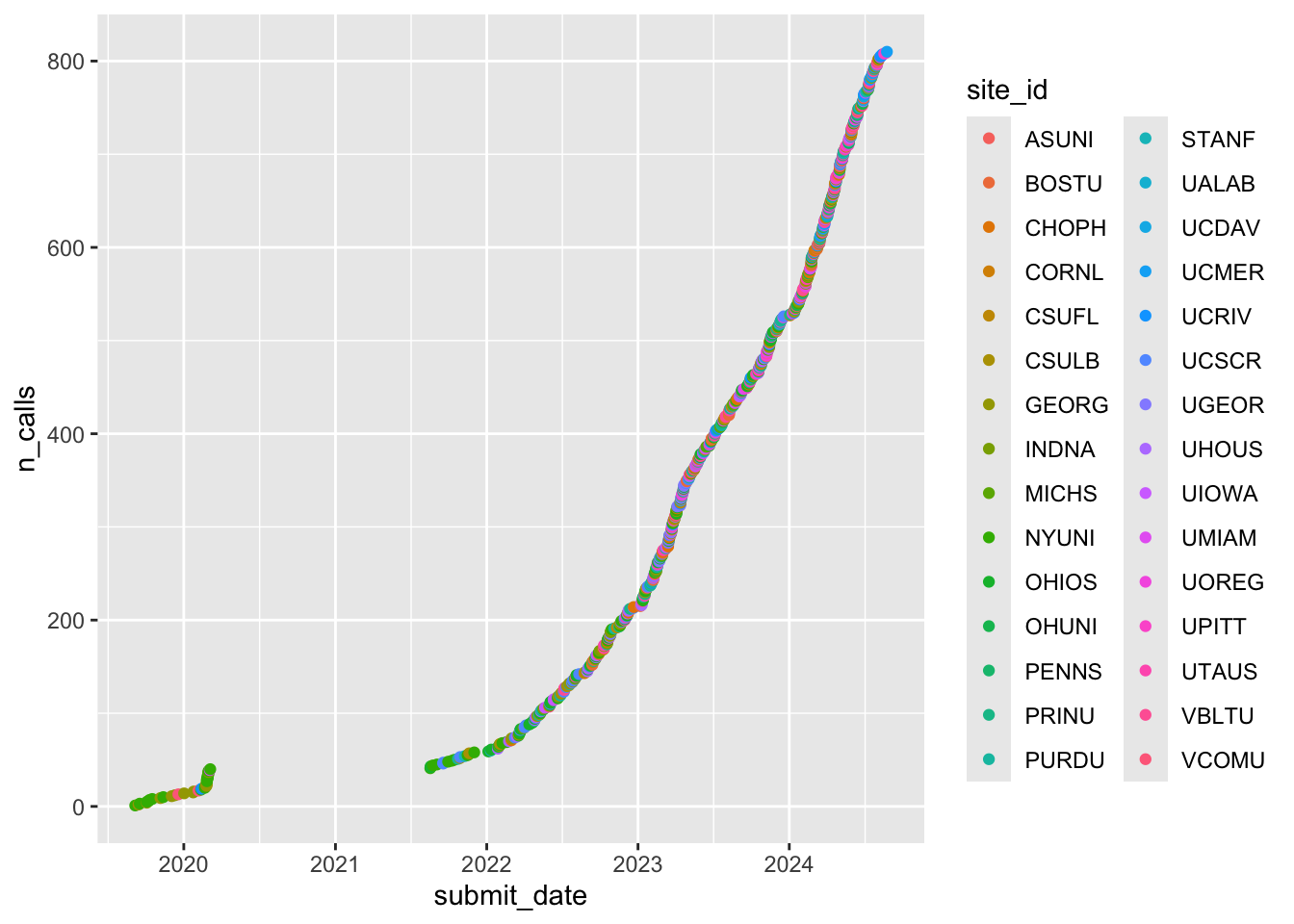
Figure 1: Cumulative screening calls by year and site
Calls by site
calls_by_site_plot <- function(df) {
df |>
filter(!is.na(site_id)) %>%
ggplot() +
aes(fct_infreq(site_id), fill = site_id) +
geom_bar() +
theme(axis.text.x = element_text(
angle = 90,
vjust = 0.5,
hjust = 1
)) + # Rotate text
labs(x = "site") +
theme(legend.position = "none")
}
calls_by_site_plot(df)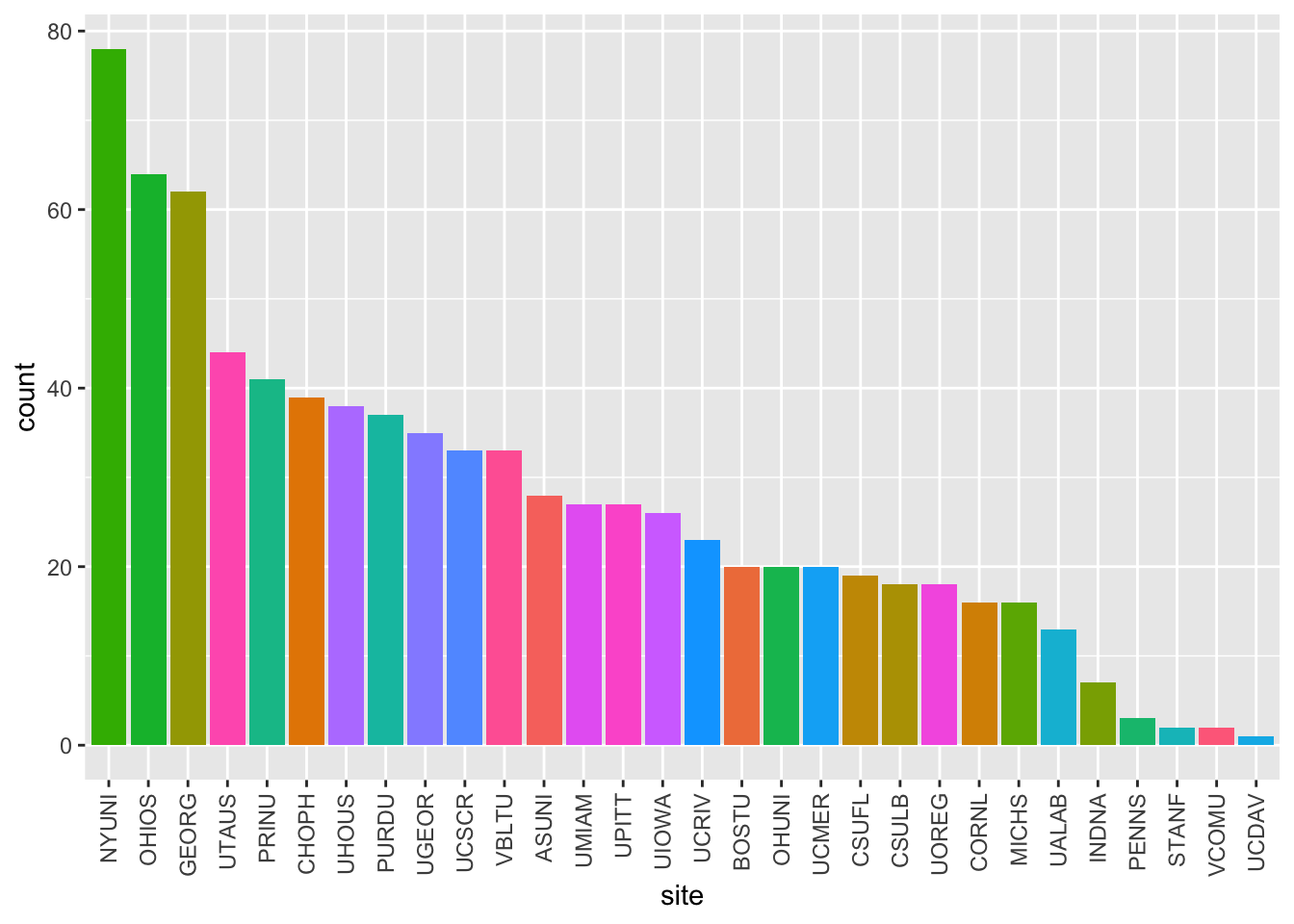
Figure 2: Cumulative screening calls by site
Demographics
Child age
Child age in months (child_age_mos) by child_sex.
screen_df |>
dplyr::filter(!is.na(child_age_mos), !is.na(child_sex)) |>
ggplot() +
aes(child_age_mos, fill = child_sex) +
geom_histogram(bins = 50)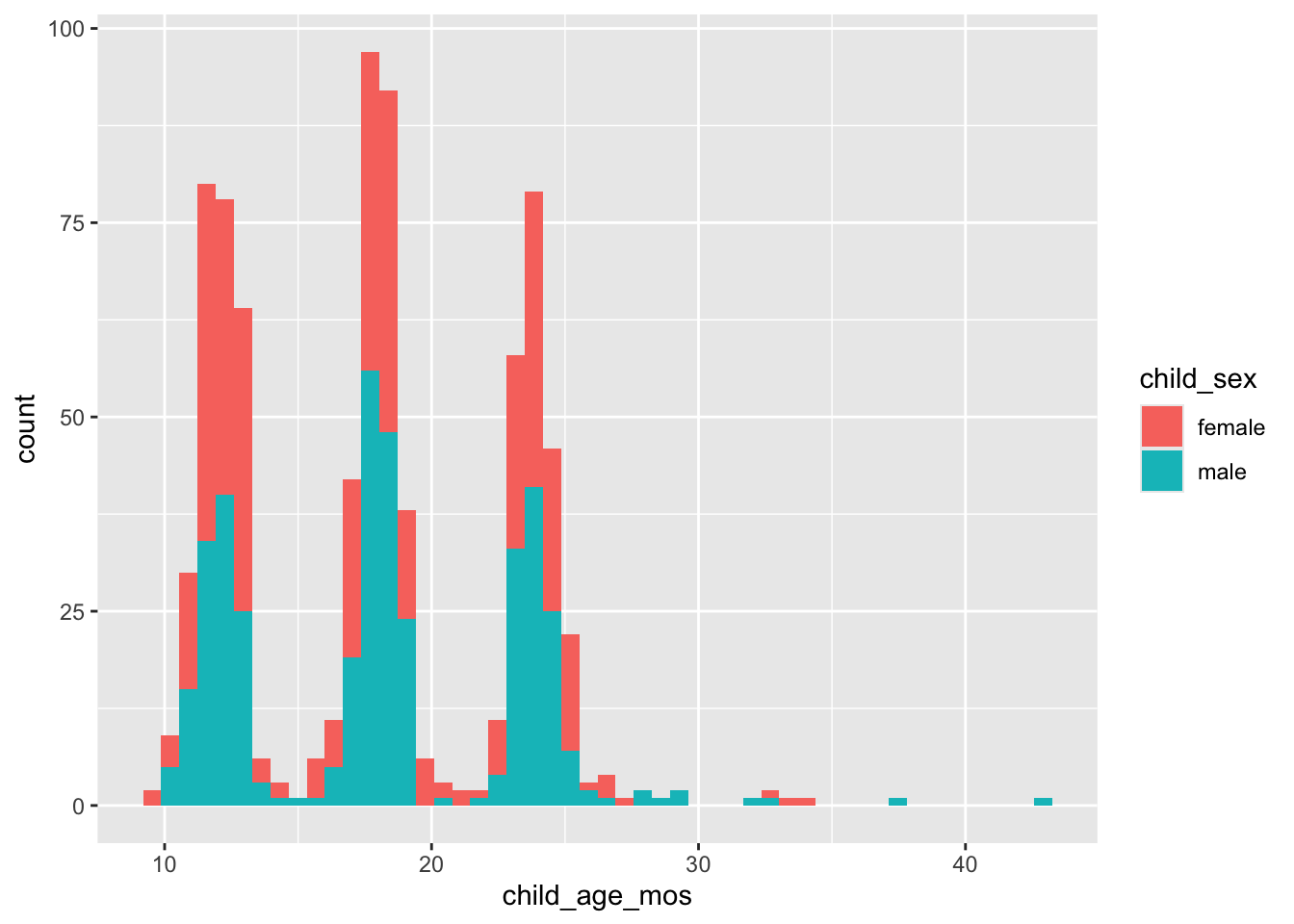
Figure 3: Histogram of child age at time of recruiting call.
Some of the code to clean the screen_df variables could be incorporated into an earlier stage of the workflow.
Language
To child
Language(s) spoken to child by child_sex.
df <- screen_df |>
dplyr::mutate(
language_spoken_child = stringr::str_replace_all(language_spoken_child, " ", "_"),
language_spoken_home = stringr::str_replace_all(language_spoken_home, " ", "_")
)
xtabs(formula = ~ child_sex + language_spoken_child,
data = df)## language_spoken_child
## child_sex english english_other english_spanish
## female 325 3 58
## male 316 5 64
## language_spoken_child
## child_sex english_spanish_other spanish
## female 3 18
## male 1 15At home
xtabs(formula = ~ child_sex + language_spoken_home, data = df)## language_spoken_home
## child_sex english english_other english_spanish
## female 331 5 47
## male 299 4 78
## language_spoken_home
## child_sex english_spanish_other other spanish
## female 1 1 18
## male 2 3 14To child vs. at home
xtabs(formula = ~ language_spoken_child + language_spoken_home, data = df)## language_spoken_home
## language_spoken_child english english_other english_spanish
## english 605 3 24
## english_other 2 6 0
## english_spanish 18 0 91
## english_spanish_other 3 0 1
## spanish 2 0 9
## language_spoken_home
## language_spoken_child english_spanish_other other spanish
## english 2 2 4
## english_other 0 0 0
## english_spanish 1 2 6
## english_spanish_other 0 0 0
## spanish 0 0 22Child health
Child born on due date
xtabs(formula = ~ child_sex + child_bornonduedate,
data = screen_df)## child_bornonduedate
## child_sex no yes
## female 8 394
## male 9 391There are \(n=\) 14 NAs.
screen_df |>
dplyr::filter(is.na(child_bornonduedate)) |>
dplyr::select(vol_id, participant_ID) |>
knitr::kable(format = 'html')| vol_id | participant_ID |
|---|---|
| 1656 | 001 |
| 1008 | 013 |
| 1008 | 013 |
| 1008 | 013 |
| 1008 | 013 |
| 1576 | 003 |
| 1576 | 006 |
| 1481 | 006 |
| 954 | 001 |
| 1103 | 001 |
| 1596 | 013 |
| 1657 | 003 |
| 979 | 026 |
| 996 | 015 |
Child term
xtabs(formula = ~ child_bornonduedate + child_onterm,
data = screen_df)## child_onterm
## child_bornonduedate no yes
## no 1 16
## yes 1 670Child weight
Must convert pounds and ounces to decimal pounds.
df <- screen_df %>%
dplyr::mutate(.,
birth_weight_lbs = child_weight_pounds + child_weight_ounces/16)
df |>
dplyr::filter(!is.na(birth_weight_lbs), !is.na(child_sex)) |>
dplyr::filter(birth_weight_lbs > 0) |>
ggplot() +
aes(x = birth_weight_lbs, fill = child_sex) +
geom_histogram(binwidth = 0.33) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())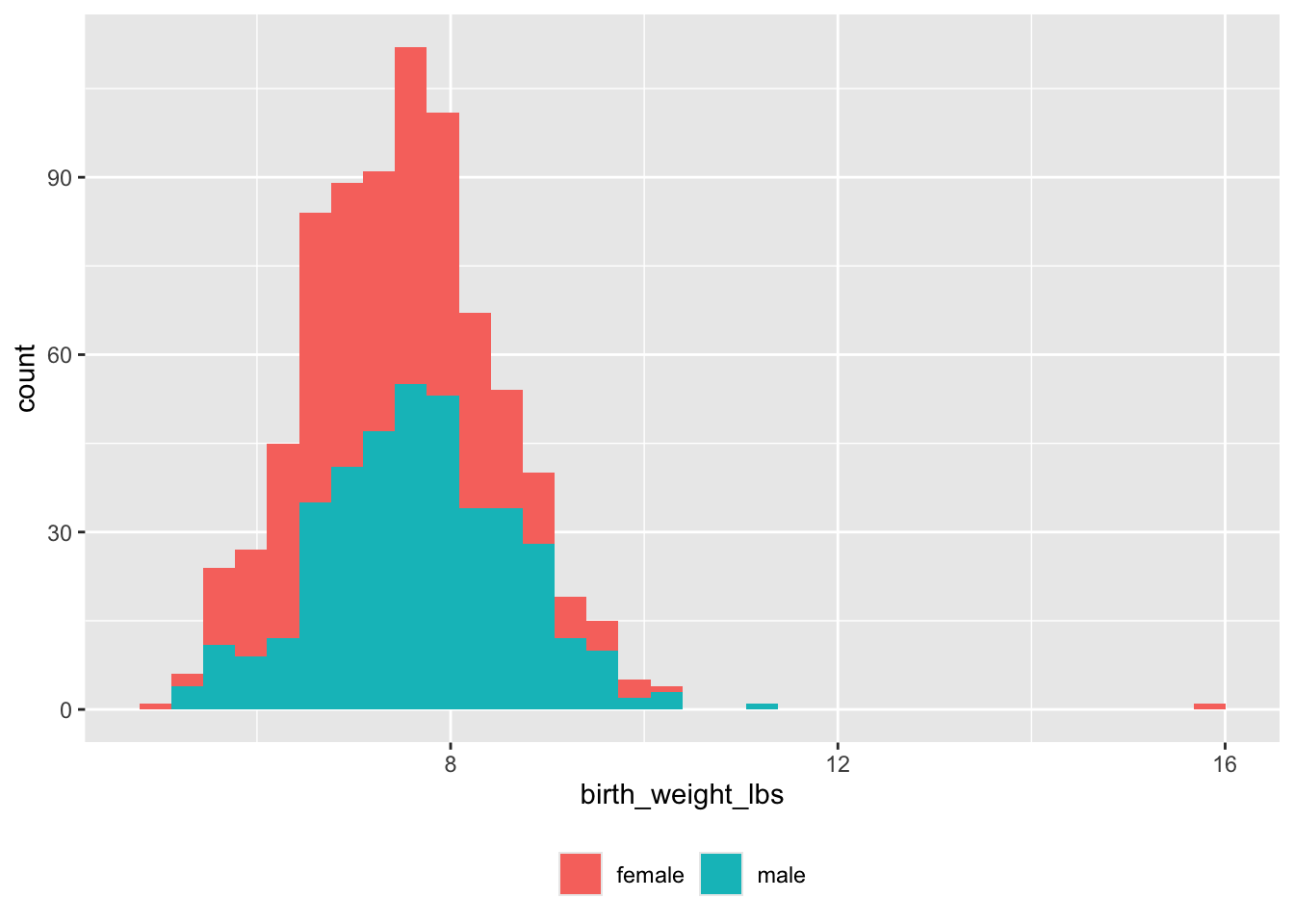
Birth complications
xtabs(formula = ~ child_sex + child_birth_complications,
data = screen_df)## child_birth_complications
## child_sex no yes
## female 363 39
## male 368 28There are some first names in the child_birth_complications_specify field, so it is not shown here.
Major illnesses or injuries
xtabs(formula = ~ child_sex + child_major_illnesses_injuries,
data = screen_df)## child_major_illnesses_injuries
## child_sex no yes
## female 389 13
## male 382 14
screen_df |>
dplyr::filter(!is.na(child_illnesses_injuries_specify),
!stringr::str_detect(child_illnesses_injuries_specify, "OK")) |>
dplyr::select(child_age_mos, child_sex, child_illnesses_injuries_specify) |>
dplyr::arrange(child_age_mos) |>
knitr::kable(format = 'html')| child_age_mos | child_sex | child_illnesses_injuries_specify |
|---|---|---|
| 11.40697 | female | COVID in January, no later complications. |
| 11.86719 | female | RSV when she 3 months; was in hospital for 2 nights |
| 12.68902 | female | fractured wrist when she was 9 months |
| 12.72189 | female | At 2 months got RSV viral infection and was in hospital for 4 days. At 5 months feel off the bed and hit her head. Ended up in the emergency room but exams showed she was okay. Neither of these events resulted in any motor, perceptual, or cognitive impairment or delay as a result. |
| 12.73285 | male | He had Covid once. |
| 12.76162 | female | RSV when 10 weeks old, hospitalized for 1 week |
| 13.01775 | male | Covid in December but this did NOT result in a visual, auditory, motor, or cognitive disability according to mom |
| 13.02186 | male | Respiratory failure and sepsis after birth that was resolved |
| 16.17494 | female | Child had a fall but she was fine. |
| 17.81860 | female | **bit by tick a couple months ago, took meds to prevent Lyme disease |
| 17.88160 | male | Covid in November |
| 18.04597 | female | broke leg in April - all healed now. slide accident, she was 16 months. in a cast for three weeks, and then a week phantom limping. |
| 18.18020 | female | Respiratory Syncytial Virus(RSV) |
| 18.67330 | male | tibial stress factor |
| 18.70617 | male | RSV but did not need to be admitted. This illness did not result in an impairment or disability. |
| 18.77055 | male | Child had COVID twice (fever, etc.) |
| 20.67856 | male | Anaphylactic reaction to peanuts |
| 22.48658 | female | Child was born with a heart murmur- cardiologist said she is unaffected and will likely grow out of it. Child also had an isolated febrile seizure short after 1st birthday. Has not experienced a seizure since. |
| 23.01118 | male | He had RSV then COVID around 13-14 months old, but they were both very mild. No complications arose from either. |
| 23.20979 | female | febrile seizure- around 1 1/2 years |
| 23.33991 | male | split lip open in March after falling on hardwood – preventative antibiotics, all clear |
| 24.03024 | male | had COVID at 7 months |
| 24.91782 | female | She had COVID, and when she was little had an cardiac issue (her heart was "working" too hard) but she was okay soon after (this was first 2-3 months after birth). March of 2021 she had fully recovered from this, and received treatment |
| 25.70814 | male | hospital day 4 for hypothermia - then negative / no diagnosis |
| 26.76008 | male | Has had a fall, was a concussion but not diagnosed with any disabilities |
| 32.67724 | male | Pneumonia at 5 months |
Child vision
xtabs(formula = ~ child_sex + child_vision_disabilities,
data = screen_df)## child_vision_disabilities
## child_sex no yes
## female 399 3
## male 394 2
screen_df |>
dplyr::filter(!is.na(child_vision_disabilities_specify)) |>
dplyr::select(child_age_mos, child_sex, child_vision_disabilities_specify) |>
dplyr::arrange(child_age_mos) |>
knitr::kable(format = 'html')| child_age_mos | child_sex | child_vision_disabilities_specify |
|---|---|---|
| 18.70617 | male | He wears glasses to correct his far sighted vision. He does not have vision loss. |
| 18.90204 | female | Duane’s Syndrome in left eye |
| 22.35371 | female | She has a lazy eye, she wears an eye patch for part of the day. |
| 23.14267 | female | Strabismus - wears glasses |
| 24.03024 | male | lazy eye but corrected |
Child hearing
xtabs(formula = ~ child_sex + child_hearing_disabilities,
data = screen_df)## child_hearing_disabilities
## child_sex no yes
## female 402 0
## male 395 1
screen_df |>
dplyr::filter(!is.na(child_hearing_disabilities_specify)) |>
dplyr::select(child_age_mos, child_sex, child_hearing_disabilities_specify) |>
dplyr::arrange(child_age_mos) |>
knitr::kable(format = 'html')| child_age_mos | child_sex | child_hearing_disabilities_specify |
|---|---|---|
| 32.67724 | male | Temporary hearing loss so tubes in his ears, one still remaining |
Child developmental delays
xtabs(formula = ~ child_sex + child_developmentaldelays,
data = screen_df)## child_developmentaldelays
## child_sex no yes
## female 336 3
## male 336 5There may be first names in the child_developmentaldelays_specify field, so it is not shown here.
Child sleep
This is work yet-to-be-done. The time stamps need to be reformatted prior to visualization.
Bed time
extract_sleep_hr <- function(t) {
t |>
stringr::str_extract("^[0-9]{2}\\:[0-9]{2}\\:[0-9]{2}") |>
hms::as_hms()
}
df <- screen_df |>
dplyr::mutate(child_sleep_time = extract_sleep_hr(child_sleep_time)) |>
dplyr::filter(!is.na(child_sleep_time))
df |>
dplyr::filter(!is.na(child_sleep_time),
!is.na(child_sex)) |>
ggplot() +
aes(child_sleep_time, fill = child_sex) +
geom_histogram(bins = 18) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())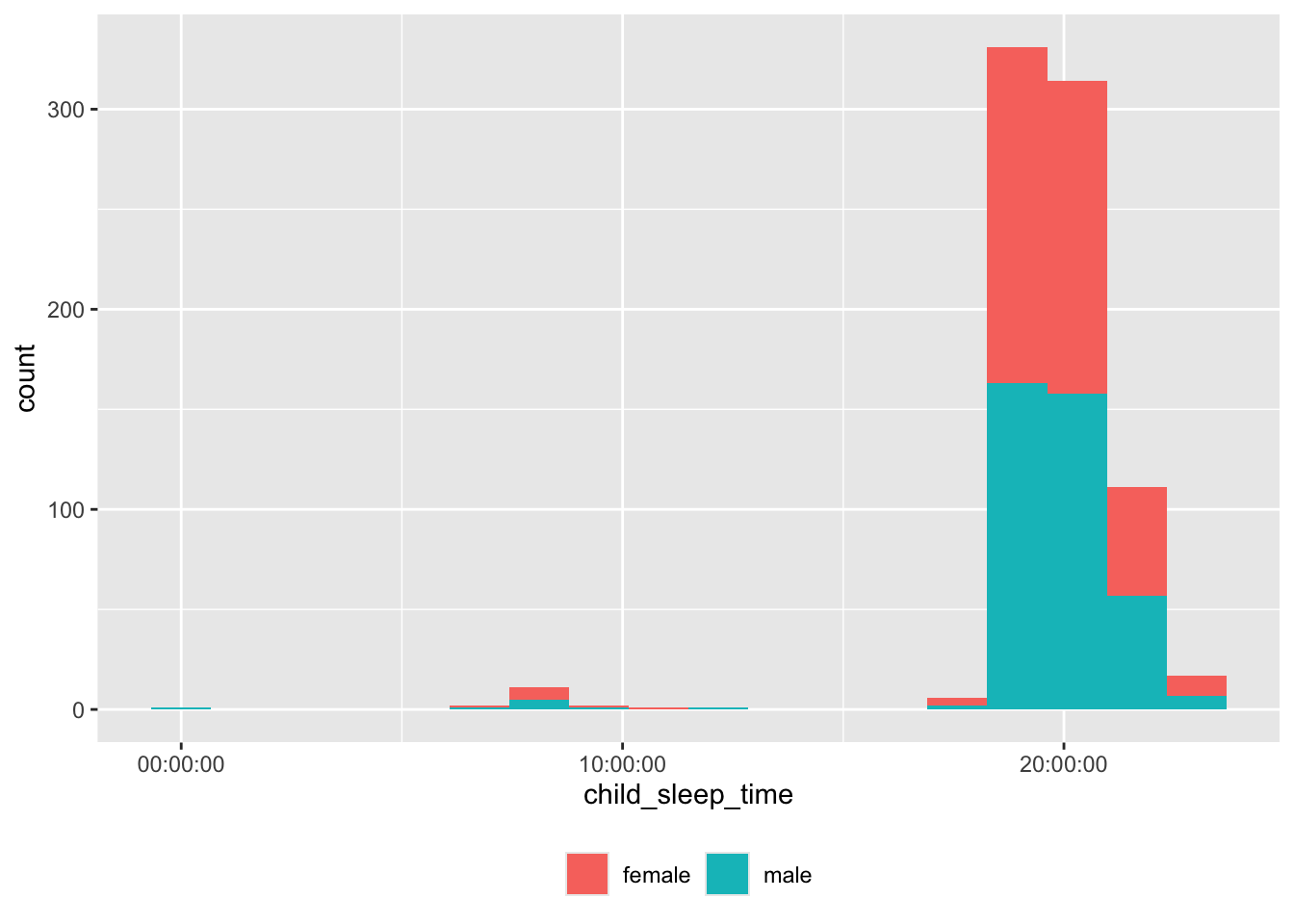
Some of the bed times are probably not in correct 24 hr time.
df |>
dplyr::filter(child_sleep_time < hms::as_hms("16:00:00")) |>
dplyr::select(site_id, participant_ID, child_sleep_time) |>
dplyr::arrange(site_id, participant_ID) |>
knitr::kable('html')| site_id | participant_ID | child_sleep_time |
|---|---|---|
| CSUFL | 013 | 07:00:00 |
| CSULB | 006 | 07:30:00 |
| INDNA | 004 | 09:00:00 |
| INDNA | 006 | 07:45:00 |
| NYUNI | 082 | 07:00:00 |
| OHIOS | 002 | 10:00:00 |
| OHUNI | 020 | 07:30:00 |
| PRINU | 001 | 12:15:00 |
| PRINU | 018 | 07:30:00 |
| PRINU | 027 | 07:30:00 |
| PRINU | 043 | 08:15:00 |
| PURDU | 020 | 08:30:00 |
| PURDU | 021 | 11:30:00 |
| UCSCR | 011 | 07:30:00 |
| UGEOR | 016 | 07:30:00 |
| UPITT | 013 | 08:15:00 |
| UPITT | 016 | 00:30:00 |
| UTAUS | 023 | 08:00:00 |
Wake time
df <- screen_df |>
dplyr::mutate(child_wake_time = extract_sleep_hr(child_wake_time)) |>
dplyr::filter(!is.na(child_wake_time))
df |>
dplyr::filter(!is.na(child_wake_time),
!is.na(child_sex)) |>
ggplot() +
aes(child_wake_time, fill = child_sex) +
geom_histogram(bins = 18) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())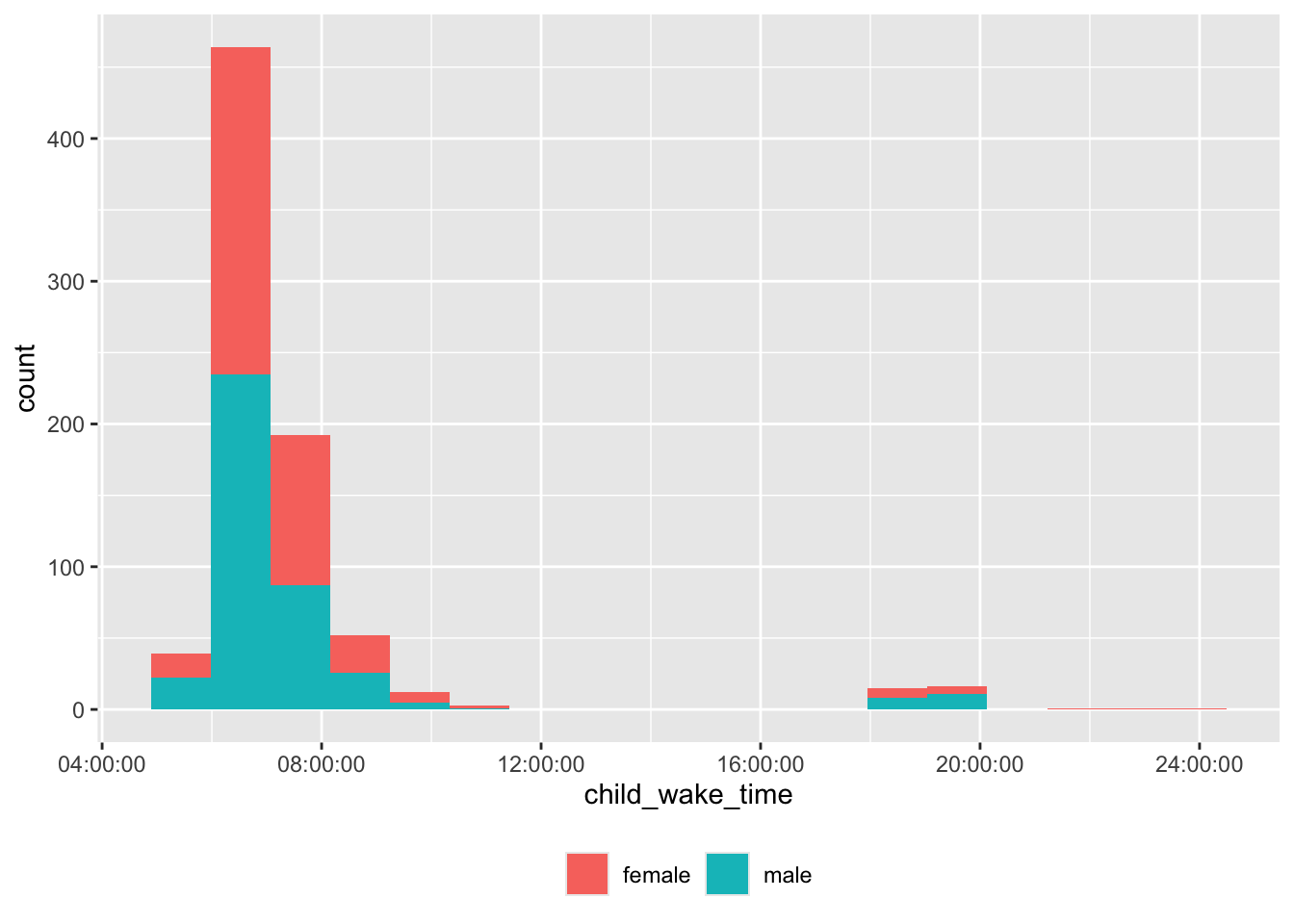
There are some unusual wake times, too.
df |>
dplyr::filter(child_wake_time > hms::as_hms("16:00:00")) |>
dplyr::select(site_id, participant_ID, child_wake_time) |>
dplyr::arrange(site_id, participant_ID) |>
knitr::kable('html')| site_id | participant_ID | child_wake_time |
|---|---|---|
| BOSTU | 001 | 19:30:00 |
| CHOPH | 017 | 19:30:00 |
| GEORG | 026 | 19:00:00 |
| OHIOS | 009 | 18:30:00 |
| OHIOS | 012 | 19:30:00 |
| OHIOS | 014 | 18:30:00 |
| OHIOS | 018 | 18:45:00 |
| OHIOS | 019 | 19:30:00 |
| OHIOS | 021 | 19:30:00 |
| OHIOS | 029 | 19:30:00 |
| OHIOS | 031 | 19:30:00 |
| OHIOS | 035 | 19:00:00 |
| OHIOS | 062 | 18:30:00 |
| PRINU | 015 | 19:00:00 |
| PRINU | 018 | 19:45:00 |
| PRINU | 027 | 20:00:00 |
| PRINU | 043 | 19:00:00 |
| PURDU | 020 | 21:30:00 |
| PURDU | 021 | 23:30:00 |
| PURDU | 023 | 23:00:00 |
| UALAB | 009 | 18:00:00 |
| UALAB | 013 | 20:00:00 |
| UCRIV | 007 | 18:00:00 |
| UCSCR | 035 | 18:30:00 |
| UGEOR | 002 | 19:30:00 |
| UHOUS | 014 | 19:00:00 |
| UMIAM | 013 | 20:00:00 |
| UMIAM | 029 | 19:30:00 |
| UPITT | 018 | 19:15:00 |
| UTAUS | 013 | 18:00:00 |
| UTAUS | 020 | 18:30:00 |
| UTAUS | 040 | 19:00:00 |
| UTAUS | 043 | 19:15:00 |
| VBLTU | 025 | 19:15:00 |
Sleep duration
df <- screen_df |>
dplyr::mutate(child_sleep_time = extract_sleep_hr(child_sleep_time),
child_wake_time = extract_sleep_hr(child_wake_time)) |>
dplyr::filter(!is.na(child_sleep_time),
!is.na(child_wake_time)) |>
dplyr::mutate(child_sleep_secs = (child_sleep_time - child_wake_time))
df |>
dplyr::filter(!is.na(child_sleep_secs)) |>
ggplot() +
aes(child_sleep_secs, fill = child_sex) +
geom_histogram(bins = 18) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())## Don't know how to automatically pick scale for object of type
## <difftime>. Defaulting to continuous.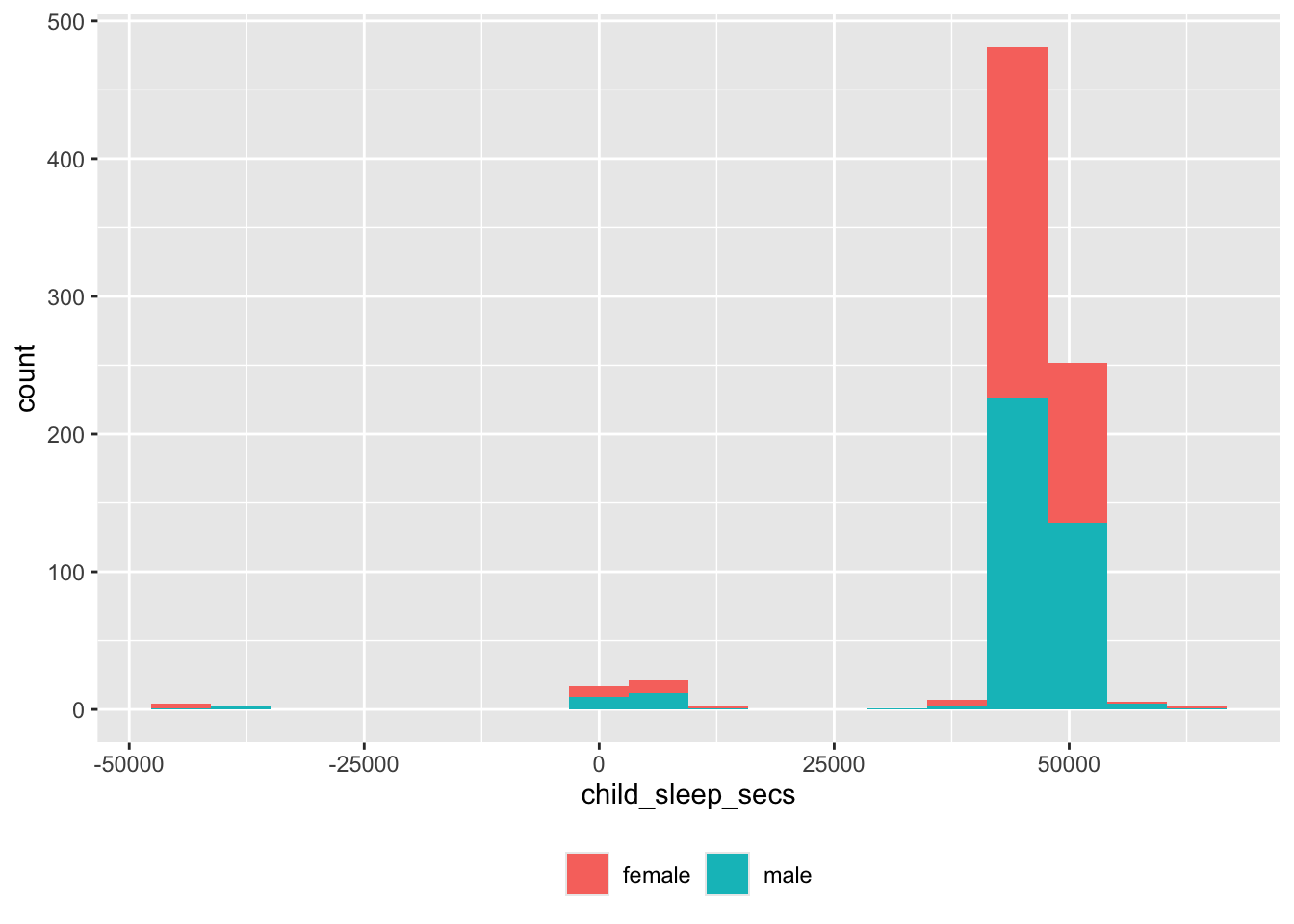
Again, there are some unusual values.
df |>
dplyr::filter(child_sleep_secs < 12000) |>
dplyr::select(site_id, participant_ID, child_sleep_time, child_wake_time, child_sleep_secs) |>
dplyr::arrange(site_id, participant_ID) |>
knitr::kable('html')| site_id | participant_ID | child_sleep_time | child_wake_time | child_sleep_secs |
|---|---|---|---|---|
| BOSTU | 001 | 19:30:00 | 19:30:00 | 0 secs |
| CHOPH | 017 | 20:45:00 | 19:30:00 | 4500 secs |
| CSUFL | 013 | 07:00:00 | 06:00:00 | 3600 secs |
| CSULB | 006 | 07:30:00 | 07:30:00 | 0 secs |
| GEORG | 026 | 19:45:00 | 19:00:00 | 2700 secs |
| INDNA | 004 | 09:00:00 | 08:00:00 | 3600 secs |
| INDNA | 006 | 07:45:00 | 06:30:00 | 4500 secs |
| NYUNI | 082 | 07:00:00 | 06:15:00 | 2700 secs |
| OHIOS | 002 | 10:00:00 | 08:00:00 | 7200 secs |
| OHIOS | 009 | 18:30:00 | 18:30:00 | 0 secs |
| OHIOS | 012 | 20:00:00 | 19:30:00 | 1800 secs |
| OHIOS | 014 | 20:00:00 | 18:30:00 | 5400 secs |
| OHIOS | 018 | 20:00:00 | 18:45:00 | 4500 secs |
| OHIOS | 019 | 20:30:00 | 19:30:00 | 3600 secs |
| OHIOS | 021 | 20:00:00 | 19:30:00 | 1800 secs |
| OHIOS | 029 | 20:45:00 | 19:30:00 | 4500 secs |
| OHIOS | 031 | 20:30:00 | 19:30:00 | 3600 secs |
| OHIOS | 035 | 20:30:00 | 19:00:00 | 5400 secs |
| OHIOS | 062 | 20:30:00 | 18:30:00 | 7200 secs |
| OHUNI | 020 | 07:30:00 | 06:00:00 | 5400 secs |
| PRINU | 015 | 19:00:00 | 19:00:00 | 0 secs |
| PRINU | 018 | 07:30:00 | 19:45:00 | -44100 secs |
| PRINU | 027 | 07:30:00 | 20:00:00 | -45000 secs |
| PRINU | 043 | 08:15:00 | 19:00:00 | -38700 secs |
| PURDU | 020 | 08:30:00 | 21:30:00 | -46800 secs |
| PURDU | 021 | 11:30:00 | 23:30:00 | -43200 secs |
| PURDU | 023 | 22:30:00 | 23:00:00 | -1800 secs |
| UALAB | 009 | 19:30:00 | 18:00:00 | 5400 secs |
| UALAB | 013 | 19:30:00 | 20:00:00 | -1800 secs |
| UCRIV | 007 | 21:00:00 | 18:00:00 | 10800 secs |
| UCSCR | 011 | 07:30:00 | 06:30:00 | 3600 secs |
| UCSCR | 035 | 20:45:00 | 18:30:00 | 8100 secs |
| UGEOR | 002 | 20:00:00 | 19:30:00 | 1800 secs |
| UGEOR | 016 | 07:30:00 | 08:00:00 | -1800 secs |
| UHOUS | 014 | 19:00:00 | 19:00:00 | 0 secs |
| UMIAM | 013 | 21:00:00 | 20:00:00 | 3600 secs |
| UMIAM | 029 | 20:00:00 | 19:30:00 | 1800 secs |
| UPITT | 013 | 08:15:00 | 07:15:00 | 3600 secs |
| UPITT | 016 | 00:30:00 | 11:00:00 | -37800 secs |
| UPITT | 018 | 19:15:00 | 19:15:00 | 0 secs |
| UTAUS | 013 | 19:30:00 | 18:00:00 | 5400 secs |
| UTAUS | 020 | 20:00:00 | 18:30:00 | 5400 secs |
| UTAUS | 023 | 08:00:00 | 07:00:00 | 3600 secs |
| UTAUS | 040 | 19:00:00 | 19:00:00 | 0 secs |
| UTAUS | 043 | 22:00:00 | 19:15:00 | 9900 secs |
| VBLTU | 025 | 19:30:00 | 19:15:00 | 900 secs |
Nap hours
df <- screen_df |>
dplyr::mutate(child_nap_hours = as.numeric(child_nap_hours)) |>
dplyr::filter(!is.na(child_sleep_time)) ## Warning: There was 1 warning in `dplyr::mutate()`.
## ℹ In argument: `child_nap_hours = as.numeric(child_nap_hours)`.
## Caused by warning:
## ! NAs introduced by coercion
df |>
dplyr::filter(!is.na(child_nap_hours),
!is.na(child_sex)) |>
ggplot() +
aes(child_nap_hours, fill = child_sex) +
geom_histogram(bins = 18) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())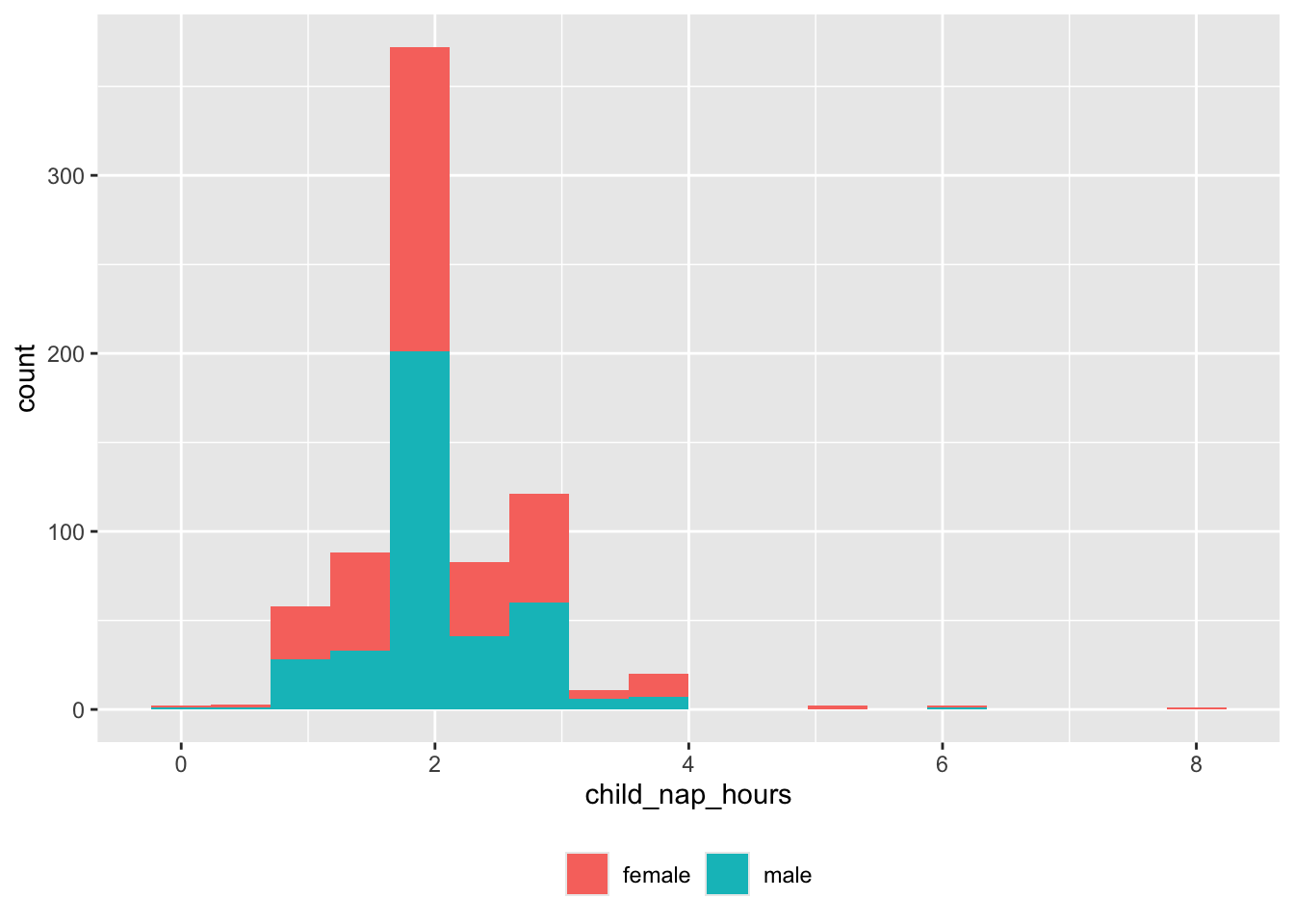
And there are some very long nappers or null values we need to capture.
df |>
dplyr::filter(child_nap_hours > 5) |>
dplyr::select(site_id, participant_ID, child_nap_hours) |>
dplyr::arrange(site_id, participant_ID) |>
knitr::kable('html')| site_id | participant_ID | child_nap_hours |
|---|---|---|
| UMIAM | 003 | 8 |
| UOREG | 012 | 6 |
| UTAUS | 009 | 6 |
Sleep location
xtabs(formula = ~ child_sex + child_sleep_location,
data = screen_df)Mother
Biological or adoptive
xtabs(formula = ~ child_sex + mom_bio,
data = screen_df)## mom_bio
## child_sex no_adoptive no_partnerchild yes
## female 2 1 385
## male 0 0 391Age at childbirth
screen_df |>
dplyr::filter(!is.na(mom_childbirth_age), !is.na(child_sex)) |>
ggplot() +
aes(x = mom_childbirth_age, fill = child_sex) +
geom_histogram(bins = 25) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())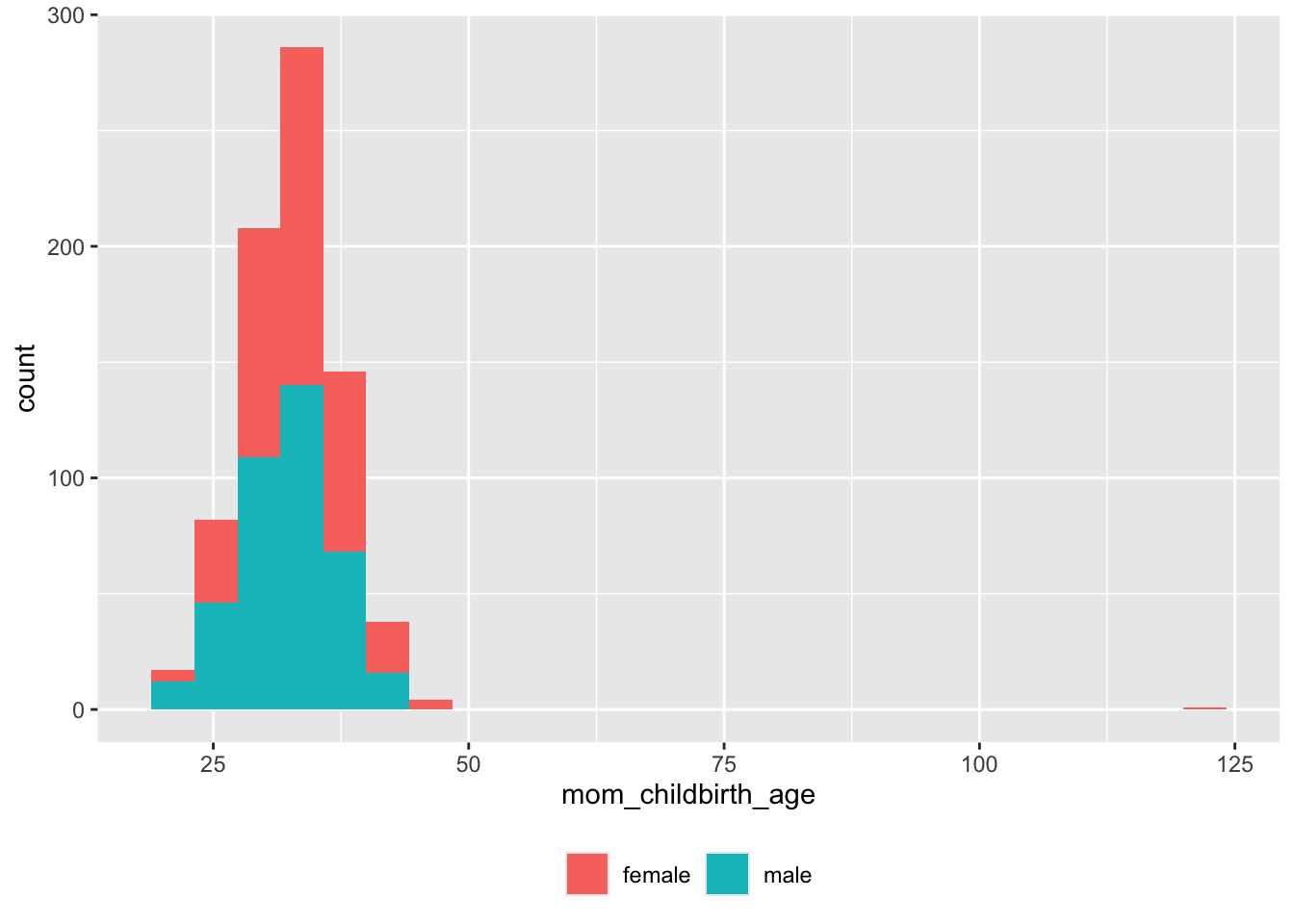
Clearly, there are some impossible (erroneous) maternal ages > 100. Here are details:
old_moms <- screen_df |>
dplyr::filter(mom_childbirth_age > 55)
old_moms |>
dplyr::select(submit_date, vol_id, participant_ID, mom_childbirth_age) |>
knitr::kable(format = 'html')| submit_date | vol_id | participant_ID | mom_childbirth_age |
|---|---|---|---|
| 2022-07-07 15:03:20 | 1391 | 005 | 121.22 |
Race
df <- screen_df |>
dplyr::filter(!is.na(mom_race)) |>
dplyr::mutate(mom_race = dplyr::recode(
mom_race,
morethanone = "more_than_one",
americanindian = "american_indian"))
xtabs(~mom_race, df)## mom_race
## american_indian asian black more_than_one
## 3 25 36 43
## nativehawaiian other refused white
## 1 48 2 634Birth country
df <- screen_df |>
dplyr::mutate(mom_birth_country = dplyr::recode(
mom_birth_country,
unitedstates = "US",
united_states = "US",
othercountry = "Other",
other_country = "Other",
puertorico = "Puerto Rico",
refused = "Refused"
))
xtabs(~ mom_birth_country + child_sex, data = df)## child_sex
## mom_birth_country female male
## Other 45 42
## Puerto Rico 1 2
## US 351 351
df <- screen_df |>
dplyr::mutate(mom_birth_country_specify =
stringr::str_to_title(mom_birth_country_specify)) |>
dplyr::filter(!is.na(mom_birth_country_specify)) |>
dplyr::mutate(mom_birth_country_specify = dplyr::recode(
mom_birth_country_specify,
Uk = "United Kingdom",
Colomobia = "Colombia",
Columbia = "Colombia"
)) |>
dplyr::arrange(mom_birth_country_specify) |>
dplyr::select(child_sex, mom_birth_country_specify)
unique(df$mom_birth_country_specify)## [1] "Argentina" "Australia"
## [3] "Bolivia" "Canada"
## [5] "Chile" "China"
## [7] "Colombia" "Costa Rica"
## [9] "Dominican Republic" "Ecuador"
## [11] "El Salvador" "England"
## [13] "Guatemala" "Haiti"
## [15] "Honduras" "India"
## [17] "Ireland" "Italy"
## [19] "Kenya" "Liberia"
## [21] "Malaysia" "Mexico"
## [23] "Nigeria" "Pakistan"
## [25] "Panama" "Peru"
## [27] "Phillipines" "Refused"
## [29] "South Korea" "Spain"
## [31] "United Kingdom" "Venezuela"
## [33] "Zimbabwe"
xtabs(~ mom_birth_country_specify + child_sex , data = df)## child_sex
## mom_birth_country_specify female male
## Argentina 1 0
## Australia 1 1
## Bolivia 0 1
## Canada 6 3
## Chile 2 0
## China 0 1
## Colombia 5 1
## Costa Rica 0 1
## Dominican Republic 0 1
## Ecuador 2 1
## El Salvador 1 1
## England 3 0
## Guatemala 0 2
## Haiti 0 1
## Honduras 1 0
## India 2 0
## Ireland 1 0
## Italy 1 0
## Kenya 0 1
## Liberia 1 0
## Malaysia 0 1
## Mexico 8 14
## Nigeria 0 1
## Pakistan 1 0
## Panama 1 0
## Peru 0 1
## Phillipines 0 1
## Refused 0 1
## South Korea 1 2
## Spain 2 1
## United Kingdom 2 1
## Venezuela 3 2
## Zimbabwe 0 1Education
df <- screen_df |>
dplyr::filter(!is.na(mom_education)) |>
dplyr::select(child_sex, mom_education)
xtabs(~ mom_education, data = df)## mom_education
## associates bachelor_s_deg bachelors
## 27 3 254
## diploma doctorate eleventh
## 27 98 1
## ged graduate_nodegree master_s_degre
## 3 24 2
## masters ninth professional
## 268 1 35
## professional_d somecollege voc_diplima
## 2 42 3
## voc_nodiplima
## 1This requires some recoding work.
Occupation
This information is available, but would need to be substantially recoded to be useful in summary form.
Jobs number
## mom_jobs_number
## 0 1 2 3 4 5more
## 1 543 60 1 1 1
df <- screen_df |>
dplyr::filter(!is.na(mom_jobs_number),
!is.na(mom_employment))
xtabs(~ mom_jobs_number + mom_employment, data = df)## mom_employment
## mom_jobs_number full_time part_time
## 0 0 1
## 1 413 130
## 2 35 25
## 3 1 0
## 4 1 0
## 5more 1 0Biological father
Race
df <- screen_df |>
dplyr::select(biodad_childbirth_age, biodad_race, child_sex) |>
dplyr::mutate(
biodad_race =
dplyr::recode(
biodad_race,
americanindian_NA = "american_indian",
asian_NA = "asian",
NA_asian = "asian",
black_NA = "black",
donotknow_NA = "do_not_know",
NA_NA = "NA",
NA_white = "white",
other_NA = "other",
refused_NA = "refused",
white_NA = "white",
morethanone_NA = "more_than_one"
)
) |>
dplyr::mutate(biodad_childbirth_age = stringr::str_remove_all(biodad_childbirth_age, "[_NA]")) |>
# dplyr::filter(!is.na(biodad_childbirth_age), !is.na(biodad_race)) |>
dplyr::mutate(biodad_childbirth_age = as.numeric(biodad_childbirth_age))## Warning: There was 1 warning in `dplyr::mutate()`.
## ℹ In argument: `biodad_childbirth_age =
## as.numeric(biodad_childbirth_age)`.
## Caused by warning:
## ! NAs introduced by coercion
xtabs(~ biodad_race, df)## biodad_race
## american_indian asian black
## 4 28 43
## do_not_know more_than_one NA
## 5 35 31
## nativehawaiian_NA other refused
## 2 65 3
## white
## 594Age at child birth
df |>
dplyr::filter(!is.na(biodad_childbirth_age)) |>
ggplot() +
aes(x = biodad_childbirth_age, fill = child_sex) +
geom_histogram(bins = 25) +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())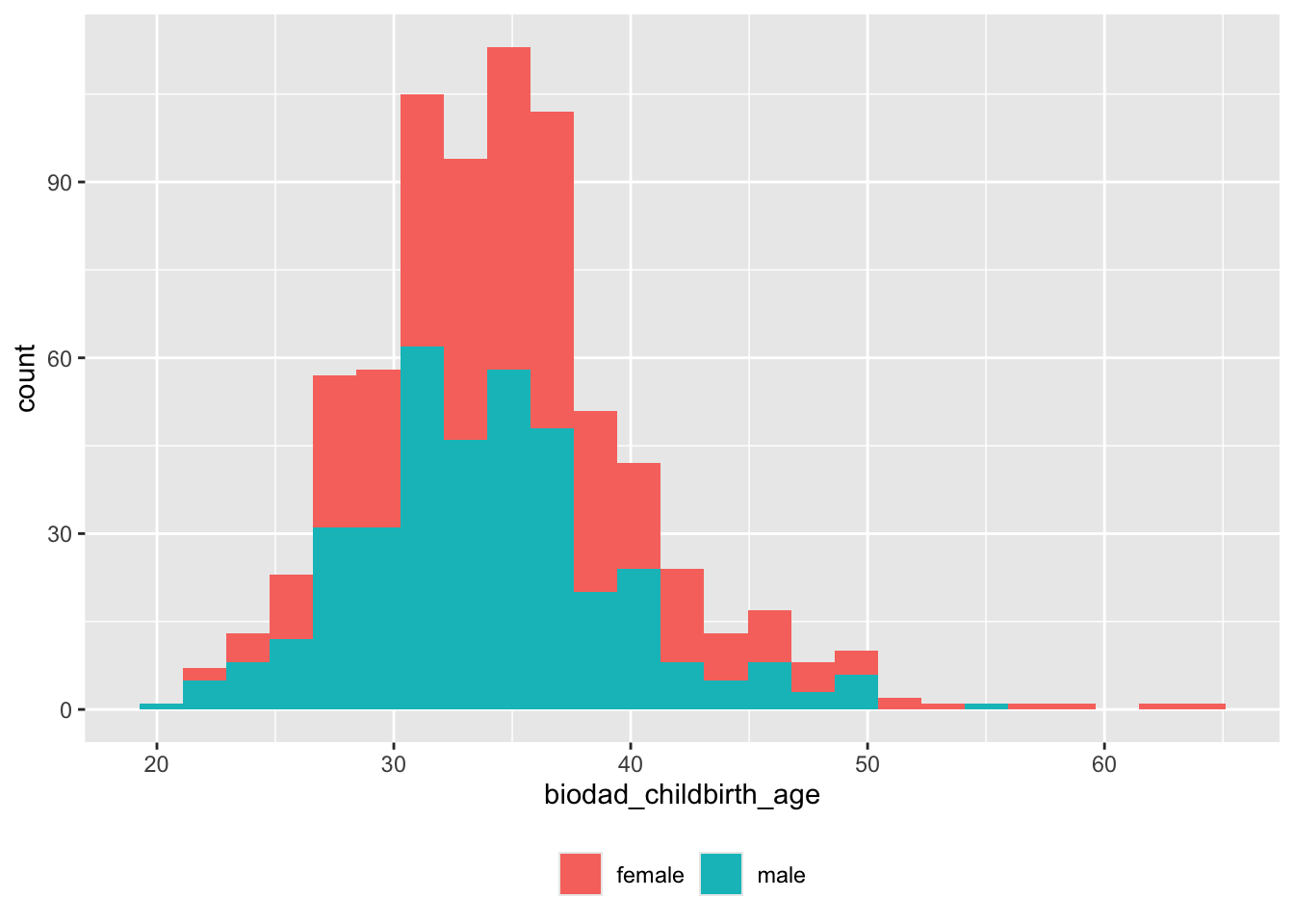
Childcare
Types
## childcare_types
## childcare
## 224
## childcare_cent
## 2
## nanny
## 4
## nanny_babysitt
## 1
## nanny_home
## 39
## nanny_home childcare
## 13
## nanny_home nanny_nothome
## 2
## nanny_home nanny_nothome relative
## 1
## nanny_home relative
## 6
## nanny_nothome
## 15
## nanny_nothome childcare
## 3
## nanny_nothome relative
## 3
## none
## 178
## relative
## 94
## relative childcare
## 19This requires some cleaning.
Hours
df <- screen_df |>
dplyr::filter(!is.na(childcare_hours)) |>
dplyr::arrange(childcare_hours)
unique(df$childcare_hours)## [1] "1"
## [2] "10"
## [3] "12"
## [4] "13"
## [5] "14"
## [6] "14.5"
## [7] "15"
## [8] "15-20"
## [9] "16"
## [10] "16-20"
## [11] "17"
## [12] "17.5"
## [13] "18"
## [14] "19"
## [15] "2"
## [16] "2-3"
## [17] "2-4"
## [18] "20"
## [19] "20 hours"
## [20] "20 hours a week - maternal grandmother"
## [21] "21"
## [22] "21 (14 hr childcare center, 7 hr babysitter)"
## [23] "22"
## [24] "24"
## [25] "25"
## [26] "25, 15 of those hours are at daycare"
## [27] "26"
## [28] "27"
## [29] "28"
## [30] "3"
## [31] "30"
## [32] "30-35"
## [33] "30-40"
## [34] "31"
## [35] "32"
## [36] "33"
## [37] "34"
## [38] "35"
## [39] "35-40"
## [40] "36"
## [41] "36; Grandpa watches one day a week"
## [42] "37"
## [43] "38"
## [44] "4"
## [45] "40"
## [46] "40+"
## [47] "40-45"
## [48] "40-45 hours"
## [49] "40-50"
## [50] "42"
## [51] "42.5"
## [52] "43"
## [53] "45"
## [54] "45-50"
## [55] "47"
## [56] "5"
## [57] "50"
## [58] "52.5"
## [59] "53"
## [60] "6"
## [61] "63"
## [62] "7"
## [63] "8"
## [64] "8 hours with grandmother"
## [65] "8-16"
## [66] "8-5 each day"
## [67] "8.5"
## [68] "80"
## [69] "9"This requires some cleaning.
Language
## [1] "Spanish and English (half and half)"
## [2] "English"
## [3] "Spanish"
## [4] "English and Spanish (50/50)"
## [5] "english"
## [6] "English when speaking with children, but Arabic when speaking to eachother"
## [7] "Mostly English, some Spanish"
## [8] "Mandarin"
## [9] "Spanish at the center"
## [10] "English and Spanish"
## [11] "English, Spanish"
## [12] "Usually English, sometimes arabic and french"
## [13] "English and speaks a little bit of Japanese but not directly to Lucas"
## [14] "English and Creole"
## [15] "Spanish (sometimes English)"
## [16] "English;Spanish"
## [17] "Spanish and English"
## [18] "spanish"
## [19] "French only"
## [20] "English and some sign language"
## [21] "Engish"
## [22] "English, American Sign Language"
## [23] "English and ASL"
## [24] "Ingles"
## [25] "English/Spanish"
## [26] "English and some Spanish"
## [27] "english, has been exposed to Arabic"
## [28] "Nanny in Spanish at home. At school both English and Spanish."
## [29] "English (majority of the time) and sometimes Spanish"
## [30] "English and Spanish 50/50"
## [31] "english, some spanish"
## [32] "English, Italian, Spanish"
## [33] "English, sometimes a little bit of Spanish"
## [34] "English & Spanish"
## [35] "English and Portuguese"This requires some cleaning.